Abstract
This report describes SARS-CoV-2 genomic surveillance conducted by the Department of Defense (DoD) Global Emerging Infections Surveillance Branch and the Next-Generation Sequencing and Bioinformatics Consortium (NGSBC) in response to the COVID-19 pandemic. Samples and sequence data were from SARS-CoV-2 infections occurring among Military Health System (MHS) beneficiaries from 1 March to 31 December 2020. There were 1,366 MHS samples sequenced from 10 countries, 36 U.S states or territories, and 5 Geographic Combatant Commands, representing approximately 2% of DoD cases in 2020. Genomes from these samples were compared with other public sequences; observed trends were similar to those of Centers for Disease Control and Prevention national surveillance in the U.S. with B.1, B.1.2, and other sub-lineages comprising the dominant variants of SARS-CoV-2. Sequence data were used to monitor transmission dynamics on U.S. Navy ships and at military training centers and installations. As new variants emerge, DoD medical and public health practitioners should maximize the use of genomic surveillance resources within DoD to inform force health protection measures.
What are the new findings?
From 1 March through 31 December 2020, 1,366 MHS SARS-CoV-2 sequences were generated from 36 U.S. states or territories, 10 countries, and 9 naval vessels. Dominant lineages detected were B.1.2 (17%), B.1 (14%) and B.1.1 (8%). The first MHS case of a variant of concern (Epsilon) was identified in December 2020.
What is the impact on readiness and force health protection?
As demonstrated by the emergence of multiple SARS-CoV-2 variants, sequencing is needed to monitor viral evolution and inform mitigation strategies in DoD settings, including troop movement and hygiene measures. Genomic surveillance supports the development of diagnostics, therapeutics, and vaccines and can help monitor their effectiveness over time.
Background
In 2020, approximately 138,000 probable COVID-19 cases, including more than 87,000 polymerase chain reaction-(PCR)-confirmed cases, were reported among U.S. service members, dependents, and retirees (Shauna Stahlman, PhD, email communication, December 2021). During the early pandemic, SARS-CoV-2 transmission had dramatic impacts on Department of Defense (DoD) operations in the U.S. and overseas, with outbreaks limiting personnel movement in South Korea, sidelining U.S. Navy ships, and curtailing training on installations.1-3 Significant time and resources were required for COVID-19 surveillance and mitigation of its impacts on DoD operations and Military Health System (MHS) beneficiaries.
COVID-19 is caused by SARS-CoV-2, which is a member of family Coronaviridae, genus Beta coronavirus.
The virus contains a roughly 30 kilobase positive-sense RNA genome encoding 4 structural and 16 non-structural viral proteins.
Since the beginning of the pandemic, viral sequencing has been a critical component of the response.4-6 The first sequenced and publicly released SARS-CoV-2 genomes helped guide development of diagnostic assays and eventually vaccines.7 Genomic data have also become critical components of epidemiology and outbreak response.
These data can provide information about large-scale epidemics, such as the approximate date when SARS-CoV-2 was first introduced to North America,8 and can also be used to characterize localized outbreaks, such as in congregate living facilities9 and on cruise ships.10 SARS-CoV-2 genomic epidemiology has also been used to examine transmission events within military recruit and trainee settings11 and to detect introductions of novel variants from international military deployments.12
Genomic surveillance using wholegenome sequencing (WGS) is an important tool that can be used to detect changes in the SARS-CoV-2 viral genome. These changes may affect diagnostic sensitivity, vaccine efficacy, monoclonal antibody efficacy, or viral transmission and virulence.13 More recently, the impacts of emerging SARSCoV-2 variants, collectively known as variants of concern (VOC) and variants being monitored (VBM), have been observed with respect to the effectiveness of medical countermeasures and, in the case of Alpha, Delta, and Omicron, with respect to displacement of other variants.14,15
To establish SARS-CoV-2 sequencing and genomic surveillance capabilities for the DoD, the Armed Forces Health Surveillance Division (AFHSD) Global Emerging Infections Surveillance (GEIS) Branch leveraged existing partnerships with Army, Navy, and Air Force public health and medical research laboratories as part of the Next Generation Sequencing and Bioinformatics Consortium (NGSBC).
This Consortium was established in 2017 to work with GEIS partner DoD medical research and public health laboratories to coordinate and improve pathogen sequencing and analysis efforts. The initial efforts of the NGSBC laid a foundation for DoD expansion of WGS in 2021 in response to novel SARS-CoV-2 VOCs, to aid in investigating outbreaks and in monitoring diagnostics and vaccine effectiveness.
This report describes SARS-CoV-2 genomic surveillance findings among DoD beneficiaries from 1 March 2020 through 31 December 2020 and highlights the utility of SARS-CoV-2 sequence data for providing a baseline for further characterization of emerging variants that could impact DoD beneficiaries and operations.
Methods
In early 2020, GEIS and NGSBC representatives from U.S. Naval Medical Research Center (NMRC), U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID), Walter Reed Army Institute of Research (WRAIR), U.S. Air Force School of Aerospace Medicine (USAFSAM), U.S. Naval Health Research Center (NHRC), and overseas partner laboratories worked to rapidly evaluate protocols and establish best practices and guidance for SARS-CoV-2 WGS and standardize genomic and epidemiologic meta-data during early 2020.16 These protocols were shared with DoD laboratories in the U.S. and abroad (https://carepoint.health.mil/sites/AFHSB/geis/programmatics/ngsbc/).
Sample Collection and Selection
Samples from SARS-CoV-2 positive specimens were obtained through clinical care, outbreak response, or participation in the DoD respiratory surveillance program3 as part of the GEIS NGSBC efforts to provide advanced testing for SARS-CoV-2 (including viral isolation and WGS). Nasopharyngeal, oropharyngeal, nasal, and buccal swabs were collected and stored in universal or viral transport media. Generally, samples were collected, transported, and stored consistent with Centers for Disease Control and Prevention (CDC) interim guidance.17 Samples were selected for sequencing from clinical specimens submitted from diverse geographic locations among MHS COVID-19 cases. Samples with higher viral loads, as approximated from real-time reverse transcription polymerase chain reaction (RT-PCR) cyclethreshold (Ct) values ≤30, were prioritized.
Consistent with 45 CFR 46 guidelines, sequencing sites obtained an Institutional Review Board determination of nonhuman subject research, or a determination that sequencing was intended as a public health surveillance activity.
Laboratory Testing
Sequencing
After samples were collected and RNA was extracted, SARSCoV-2 sequencing was performed using 2 different methods (Figure 1). For the majority of samples, amplicon-based approaches were used, including the ARTIC Network and YouSeq protocols (Figure 1).18,19 Briefly, total ribonucleic acid (RNA) was extracted and reverse-transcribed into complementary deoxynucleic acid (cDNA), followed by the selective amplification of SARSCoV-2-derived cDNA using specific PCR primers. Alternatively, in some instances, to enrich for SARS-CoV-2 cDNA, hybridization approaches were applied using the Illumina Respiratory Virus Oligos Panel (Figure 1). Each sample was then barcoded and combined and sequenced in multiplexed reactions, primarily using Illumina sequencing platforms (MiSeq and NextSeq).
Analysis
As previously described, for quality control steps, low quality genome sequence reads and artifacts from sequence library preparation were removed.20,21 Individual sequence reads were then aligned to the Wuhan-Hu-1 SARS-CoV-2 reference genome (NC_045512.2).22 Consensus genomes were generated23 and classified into lineages using the PANGO (Phylogenetic Assignment of Named Global Outbreak lineage) nomenclature 24,25 and clades using Nextstrain.26,27 After clearance for public release, consensus genomes were deposited into public databases including GISAID or Genbank (https://www.ncbi.nlm.nih.gov/genbank/). Accompanying sample epidemiologic data were compiled and descriptive statistical analysis was performed using SAS/STAT software, version 9.4 (2014, SAS Institute, Cary, NC).
Results
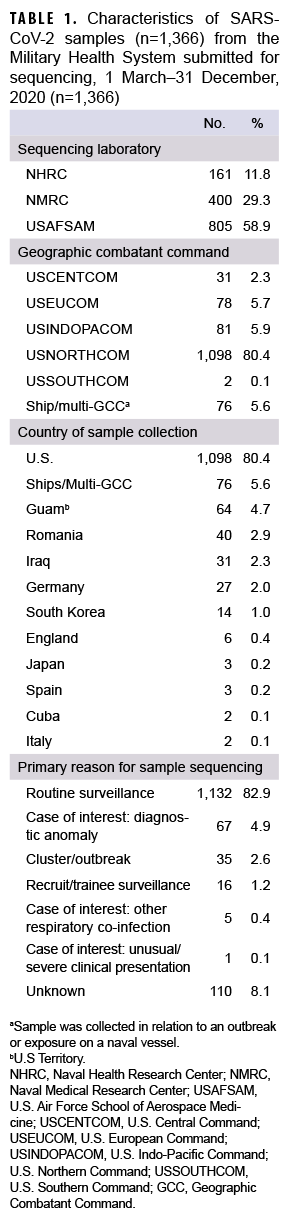
A total of 1,366 SARS-CoV-2 samples collected during 1 March to 31 December 2020 from 36 U.S. states or territories and 10 countries were sent for sequencing at NGSBC laboratories. These represented samples from individual MHS cases of COVID-19 illness and from MHS beneficiaries with preclinical/asymptomatic SARS-CoV-2 infections, collected from at least 80 military installations or military treatment facilities and 9 naval vessels within 5 Global Combatant Commands (GCC) (USEUCOM, USCENTCOM, USINDOPACOM, USNORTHCOM, and USSOUTHCOM).
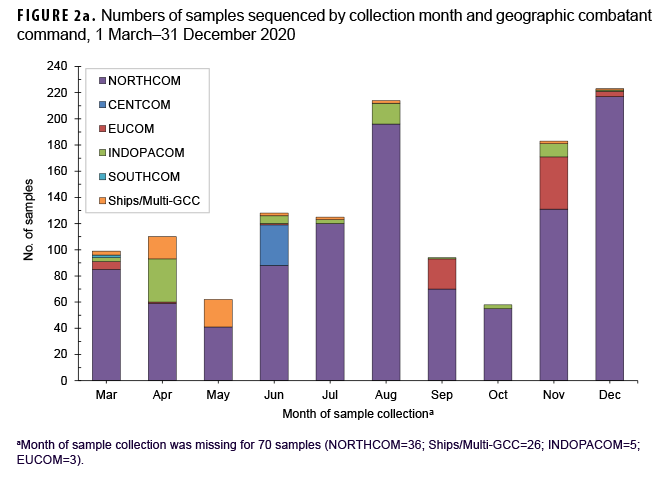
Most samples were collected in NORTHCOM (80.4%) while the remainder came from locations outside of the continental U.S. (14.0%) or naval vessels (5.6%) (Table 1). In December 2020, the 223 samples that were sequenced represented the largest monthly count of the year (Figure 2a). The second highest monthly count of samples sequenced was in August 2020 (n=214).
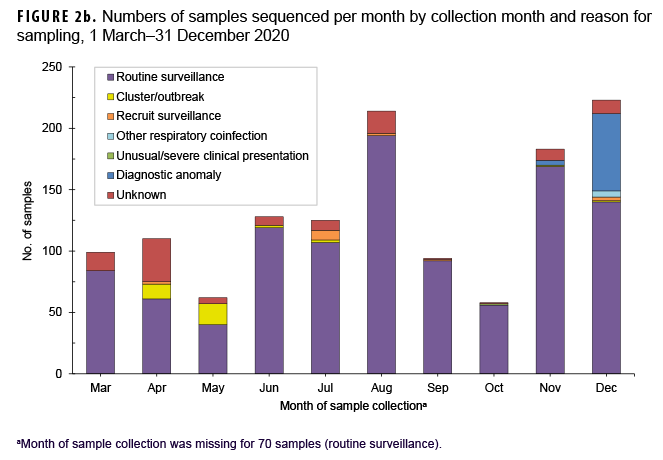
The primary reasons for submitting a sample for sequencing were routine surveillance (82.9%; n=1,132) followed by potential diagnostic anomalies (4.9%; n=67), and a suspected outbreak cluster (2.6%; n=35) (Table 1, Figure 2b). The majority of routine surveillance samples were collected as part of previously existing DoD respiratory surveillance programs or residual diagnostic and surveillance specimens. Furthermore, across all sample collection months, routine surveillance was cited as the primary reason for sequencing for the majority of samples (Figure 2b).
However, samples sent for sequencing due to suspected diagnostic anomalies, such as dropout of 1 or more RT-PCR targets, increased dramatically at the end of the year accounting for 28% (n=63) of samples collected in December 2020 (n=223) (Figure 2b).28 Most of the reported outbreak-associated samples came from public health investigations in the early half of the year; however, additional outbreak-associated clusters were identified after genomic analyses.
Sequencing results
Samples varied in terms of viral load and nucleic acid quality, and analysis of a subset of sequenced samples showed that lower RT-PCR Ct values correlated with a higher sequencing success rate.
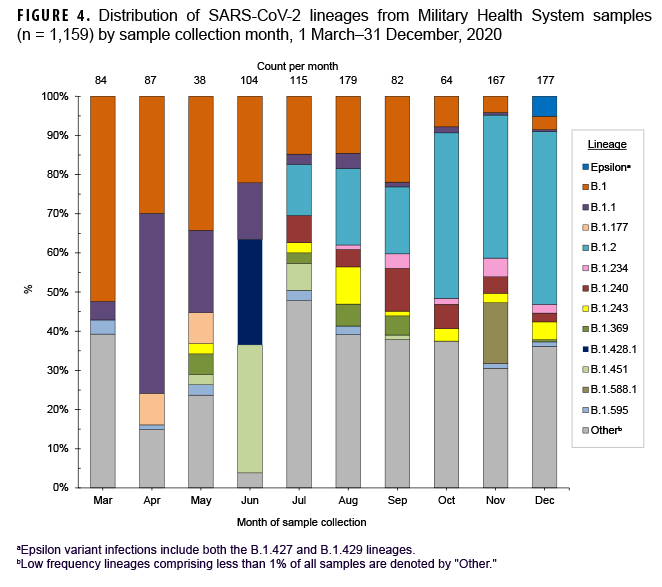
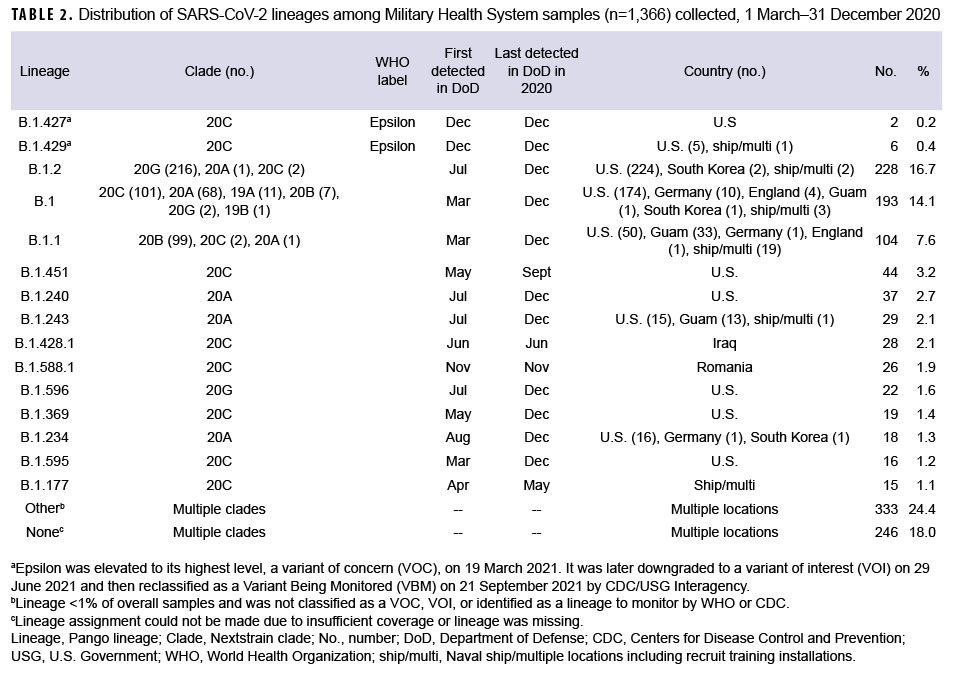
These data also showed that a Ct value of 30 or lower was a reasonable threshold for prioritizing samples for sequencing (Figure 3). Of all sequenced samples, 1,159 (84.8%) had sufficient sequence quality (i.e., breadth and depth of coverage across the genome) to generate a PANGO lineage call. The most common SARS-CoV-2 lineages in 2020 were B.1.2 (16.7%), B.1 (14.1%), and B.1.1 (7.6%).
Ten additional lineages were observed less frequently at nearly 1% each (Table 2). No single lineage was dominant in 2020; in most months, the combination of other (<1% frequency) lineages accounted for over one-third of all sequenced samples (Figure 4).
Outbreaks and suspected clusters
A subset of sequenced samples was collected from suspected outbreaks at overseas locations, on naval vessels, or at recruit training installations. Sequencing results were either used to complement more traditional epidemiological data to better describe transmission patterns or to retrospectively confirm outbreak clusters.11 Several examples include: 1) a cluster of B.1.428.1 cases (n=28) identified in samples from Iraq collected in June 2020; 2) a cluster of B.1.588.1 cases (n=26) from Romania collected in November 2020; 3) a likely outbreak of B.1.177 cases (n=10) collected from 28 April through 13 May 2020 on a Navy ship; and 4) other potential outbreaks on naval vessels and at recruit training installations (e.g., MCRD Parris Island, SC and MCRD San Diego, CA) (Table 2).
Early detection of the Epsilon variant in MHS
In December 2020, 8 Epsilon (a WHO named variant comprised of the B.1.427 and B.1.429 lineages) cases were identified in the MHS (Table 2). Two cases were infections caused by the B.1.427 lineage, identified in CA (Port Hueneme) and in OH (Wright Patterson Air Force Base (AFB). Six cases caused by the B.1.429 lineage were identified in CA (Edwards AFB and Port Hueneme), ND (Minot AFB), and from a U.S. naval vessel. Epsilon samples contained the L452R amino acid substitution in the receptor-binding domain of the spike protein associated with immune evasion and increased infectivity.29 No other WHO named variant infections were detected among the MHS samples collected and sequenced in 2020.
Editorial Comment
To respond to the emergence of SARSCoV-2, the GEIS NGSBC and other DoD partners, using existing baseline funding and resources, rapidly established sequencing and genomic surveillance for SARSCoV-2 within the MHS in early 2020. However, due to the lack of coordinated national response and DoD policy throughout 2020, less than 2% of COVID-19 cases reported within the MHS were sequenced by GEIS partners for surveillance purposes. While not all samples would have been eligible for sequencing, this demonstrates an untapped opportunity to leverage SARS-CoV-2 sequencing as an integral part of early DoD COVID-19 surveillance and response activities.
In 2020, significant barriers limited the ability of GEIS partners to obtain and sequence samples, most notably lack of awareness and consensus about privacy and regulatory requirements for sharing of samples and their associated epidemiological data during the pandemic. Similar to civilian public health settings, compliance regarding sample and data sharing across the DoD contributes to slow approval processes, delaying important analyses and reducing the timeliness of results.30
Clarity on these regulations within the DoD during a public health emergency is critical to facilitate prompt genomic surveillance priorities. It was the emergence of the Alpha (B.1.1.7) variant in the U.K. in late 2020 and its eventual global spread that generated momentum within DoD to establish a policy to recognize WGS as a critical capability in pandemic response and to expand genomic sequencing surveillance across the MHS.31
Along with efforts to characterize the clinical and public health impacts of SARSCoV-2, genomic surveillance helped guide intervention and mitigation strategies to protect U.S. service members and other MHS beneficiaries. The findings of this report, based on global genetic sequence data from MHS beneficiaries, are consistent with other reports of global and North American SARS-CoV-2 genetic diversity in 2020.32
Sequencing data also indicated that early diagnostics and countermeasures, such as vaccines and monoclonal antibodies, against the original Wuhan-Hu-1 reference would have been effective against the variants of SARS-CoV-2 circulating in the MHS in 2020. These initial surveillance efforts also provided critical baseline information and scientific infrastructure for monitoring emerging VOCs (e.g., Alpha, Beta, Gamma, Delta, Epsilon, and Omicron) and COVID-19 vaccine effectiveness in 2021.
The flexibility and robustness of this genomic surveillance program was also evident in several ways. First, as information became available about the sequencing success rate in relation to diagnostic Ct values or specific sequencing protocols, recommendations were made for prioritization of samples for sequencing and adjustments to preferred protocols. Second, in late 2020, there was a dramatic increase in the number of samples being sent for sequencing that had suspected diagnostic anomalies.
This increase was likely due to clinical and laboratory awareness of the need for enhanced national surveillance for Spike-gene target failures that could be indicative of B.1.1.7 (the variant first identified in the UK and later renamed to VOC Alpha). Finally, the geographic coverage of samples submitted from routine surveillance and outbreak response allowed for the detection of unique variants from several overseas locations in CENTCOM and EUCOM and the detection of Epsilon cases within the MHS in NORTHCOM in December 2020 (Table 2). Epsilon, first detected in southern California, was ultimately elevated to its highest level, a VOC, by the U.S. SARS-CoV-2 Interagency Group from 19 March to 29 June 2021.
There are several important limitations to the DoD WGS approach. Samples sent to NGSBC laboratories for genomic surveillance were not explicitly selected to be representative of all COVID-19 cases within the MHS but were the result of either participation in existing DoD respiratory surveillance networks or the awareness and willingness of a provider, clinician, or public health authority to share samples. Sampling bias can complicate prevalence estimates and interpretation of sequence data and missing epidemiologic data can further limit insights from phylogenetic analysis.33Finally, there is a need for better methods to account for over, under, and biased sampling for both global and regional public genomic datasets and within local outbreaks. Notably, these efforts provided some of the only SARS-CoV-2 sequence data available from locations such as Cuba, Iraq, and Romania in 2020.
A second challenge was the misunderstanding of sequencing capabilities within DoD line, medical, and public health organizations. Officials from these institutions are often too heavily burdened with frontline activities (e.g., patient care, contact tracing) to engage in what might be perceived as research. Education about the importance of DoD sequencing capabilities and genomic surveillance in the context of public health is important and can facilitate earlier access to samples and associated epidemiologic data.
Several SARS-CoV-2 VOCs have emerged in 2021 that exhibited enhanced transmission, increased severity, reduced antiviral or immunotherapy efficacy, or evasion of molecular detection assays.15 The capabilities established through this collaborative DoD effort will continue to provide critical information about SARSCoV-2 transmission patterns and help monitor medical countermeasure effectiveness for 1.4 million active component and 331,000 reserve personnel and other beneficiaries of the MHS (e.g., family members, retirees, etc.). Sequence data are also relevant for understanding transmission patterns within the DoD since military personnel are highly mobile, share close quarter accommodations during deployments, and operate globally. In an outbreak setting, sequence data can help determine if cases may result from infections acquired from local transmission while deployed or reflect transmission chains initiated prior to deployment.
In early 2020, lack of sustained funding, limitations in public health laboratory and bioinformatics infrastructure, including limited trained personnel and lack of validated protocols, impeded rapid deployment of sequencing and bioinformatics analysis for global COVID-19 response and within the civilian sector in the U.S.434
The percentage of COVID-19 cases sequenced was also far lower than several European countries with long-standing investments in genomic surveillance for infectious diseases of public health concern.35, 36 Fortunately, in the years before 2020, the GEIS NGSBC recognized the need to coordinate and standardize prior investments by DoD in infectious disease surveillance and genomic sequencing which allowed for early contributions to DoD and the interagency response during the COVID-19 pandemic.
Findings from this collaborative effort illustrate the importance of leveraging existing DoD sequencing assets and surveillance networks to rapidly respond to the COVID-19 pandemic and establish a robust global capacity to monitor SARSCoV-2 evolution in support of DoD activities. DoD should prioritize expanding genomic surveillance systems and capabilities for future pandemic preparedness.
Author affiliations
Cherokee Nation Strategic Programs, Tulsa, OK (Ms. Morton, Dr. Creppage); Defense Health Agency, Armed Forces Health Surveillance Division, Silver Spring, MD (Ms. Morton, Dr. Bazaco, Dr. Creppage, CAPT Pimentel, CAPT Scheckelhoff, LCDR Forshey, MAJ Bogue, LTC Taylor); General Dynamics Information Technology, Falls Church, VA (Dr. Bazaco); U.S. Air Force School of Aerospace Medicine, Dayton, OH (Dr. Fries, Dr. Starr, Mr. Gruner); JYG Innovations LLC, Dayton, OH (Mr. Gruner); U.S. Naval Medical Research Center, Fort Detrick, MD (Dr. Bishop-Lilly, Ms. Cer, Dr. Malagon); Leidos, Reston VA (Dr. Malagon); U.S. Naval Health Research Center, San Diego, CA (Dr. Myers, Dr. Underwood); Walter Reed Army Institute of Research, Silver Spring, MD (Dr. Maljkovic Berry); U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD (MAJ Kugelman).
Acknowledgements
The authors would like to thank all members of the Global Emerging Infections Surveillance Next-Generation Sequencing and Bioinformatics Consortium who contributed to SARS-CoV-2 genomic surveillance efforts, including the many laboratory staff who processed the samples and bioinformatics staff who analyzed genomic data. We acknowledge the important contributions of Naval Infectious Diseases Diagnostic Laboratory, Landstuhl Regional Medical Center, and the additional DOD medical treatment facility, clinical, public health, and laboratory staff that contributed to specimen collection, shipment, analysis, and data management. Additional thanks to CDR Mark Simons, LTC Grace Lidl, Dr. Simon Pollett, and LCDR Nathaniel Christy for their thoughtful conversations and help with establishing this capability early in the COVID-19 pandemic.
Disclaimer
This work was supported by the Armed Forces Health Surveillance Division, Global Emerging Infections Surveillance Branch (ProMIS ID: P0013_20_AH, P002_20_US, P0148_20_SD) and WUN A1417. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Air Force, Department of the Army, Department of the Navy, Department of Defense, or U.S. Government. Several of the authors are U.S. Government employees. This work was prepared as part of their official duties. Title 17 U.S.C. § 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
References
1. Malone JD. USS Theodore Roosevelt, COVID-19, and Ships: Lessons Learned. JAMA Netw Open. 2020;3(10): e2022095.
2. Payne DC, Smith-Jeffcoat SE, Nowak G, et al. SARS-CoV-2 Infections and Serologic Responses from a Sample of U.S. Navy Service Members - USS Theodore Roosevelt, April 2020. MMWR Morb Mortal Wkly Rep. 2020;69(23):714–721.
3. Chapleau RR, Christian M, Connors B, et al. Early Identification of SARS-CoV-2 Emergence in the Department of Defense via Retrospective Analysis of 2019-2020 Upper Respiratory Illness Samples. MSMR. 2021;28(6):2–5.
4. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–154.
5. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269.
6. Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733.
7. Virological.org. Holmes EC. Novel 2019 coronavirus genome--SARS-CoV-2 coronavirus. 10 January 2020. Accessed 3 January 2022. https://virological.org/t/novel-2019-coronavirus-genome/319
8. Worobey M, Pekar J, Larsen BB, et al. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370(6516):564-570.
9. MacCannell T, Batson J, Bonin B, et al. Genomic epidemiology and transmission dynamics of SARS-CoV-2 in congregate healthcare facilities in Santa Clara County, California. Clin Infect Dis. 2021.
10. Sekizuka T, Itokawa K, Kageyama T, et al. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc Natl Acad Sci U S A. 2020;117(33):20198–20201.
11. Letizia AG, Ramos I, Obla A, et al. SARS-CoV-2 Transmission among Marine Recruits during Quarantine. N Engl J Med. 2020;383(25):2407–2416.
12. Ballesteros N, Castaneda S, Munoz M, et al. Cluster characterization of SARS-CoV-2 in military personnel deployed to Egypt and subsequent introduction of B.1.1.7 and C.36 lineages to Colombia. J Travel Med. 2021;28(7):taab131.
13. Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424.
14. Lauring AS, Hodcroft EB. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA. 2021;325(6):529–531.
15. Lauring AS, Malani PN. Variants of SARSCoV-2. JAMA. 2021;jama.2021.14181.
16. DoD Establishes Collaborative Virus Genetic Sequencing Capability for COVID-19. 2020.
17. CDC. Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing.
18. Quick J. nCoV-2019 sequencing protocol v3. (LoCost) V.3; 25 August 2020. Accessed 3 January 2022. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye
19. Larson D, Brodniak SL, Voegtly LJ, et al. A Case of Early Reinfection With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2021;73(9):e2827–e2828.
20. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120.
21. Smith DR, Singh C, Green J, et al. Genomic and Virological Characterization of SARS-CoV-2 Variants in a Subset of Unvaccinated and Vaccinated U.S. Military Personnel. Front Med (Lausanne). 2021;8:836658.
22. Bushnell B. BBMAP: A Fast, Accurate, Splice-Aware Aligner. Poster presented at: 9th Annual Genomics of Energy & Environment Meeting; 17–20 March 2014; Walnut Creek, CA.
23. Grubaugh ND, Gangavarapu K, Quick J, et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(1):8.
24. Rambaut A, Holmes EC, O'Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407.
25. Rambaut A, Holmes EC, O'Toole A, et al. Addendum: A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2021;6(3):415.
26. I. A, C. R, E.B. H, R.A. N. Nextclade: clade assignment, mutation calling and quality control for viral genom. Journal of Open Source Software. 2021;6(65):3773.
27. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123.
28. Brown KA, Gubbay J, Hopkins J, et al. SGene Target Failure as a Marker of Variant B.1.1.7 Among SARS-CoV-2 Isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA. 2021;325(20):2115–2116.
29. Motozono C, Toyoda M, Zahradnik J, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124–1136.
30. Black A, MacCannell DR, Sibley TR, Bedford T. Ten recommendations for supporting open pathogen genomic analysis in public health. Nat Med. 2020;26(6):832-841.
31. Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(3):95–99.
32. Chen Z, Azman AS, Chen X, et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet. 2022;54(4):499–507.
33. Villabona-Arenas CJ, Hanage WP, Tully DC. Phylogenetic interpretation during outbreaks requires caution. Nat Microbiol. 2020;5(7):876–877.
34. National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Life Sciences; Health and Medicine Division; Board on Health Sciences Policy; Committee on Data Needs to Monitor the Evolution of SARS-CoV-2. In: Genomic Epidemiology Data Infrastructure Needs for SARS-CoV-2: Modernizing Pandemic Response Strategies. Washington,DC: National Academies Press; 2020:chap 5.
35. ECDC. ECDC strategic framework for the integration of molecular and genomic typing into European surveillance and multi-country outbreak investigations, 2019–2021. Stockholm: ECDC; 2019.
36. Furuse Y. Genomic sequencing effort for SARS-CoV-2 by country during the pandemic. Int J Infect Dis. 2021;103:305–307.